
Tuning Colloidal Reactions
Control over complex colloidal reactions with inverse designed components. Controlled disassembly of virus-like shells and, inspired by drug deliver applications, induced release of small particles trapped inside.

Simons Junior Fellow
NYU Center for Soft Matter Research
&
Flatiron Center for Computational Biology

Ella King will be an Assistant Professor of Chemical and Biological Engineering and of Materials Science and Engineering at Northwestern University beginning September 2026. Interested students and postdocs are welcome to contact her! Currently, Ella is a Simons Junior Fellow at the NYU Center for Soft Matter Research and the
Flatiron Center for Computational Biology in New York City. She received her Ph.D. in Physics from Harvard University and B.S. from Stanford University. Full CV
The King group works to understand and design materials that are dynamic, responsive, and can correct their own errors. Unlike human-made materials, biological systems are strikingly robust to errors in the face of dynamic, out-of-equilibrium environments. Living matter corrects errors across six orders of magnitude: nanometer-scale kinetic proofreading achieves exponentially lower error rates in DNA replication than can be reached at equilibrium, and millimeter-scale vasculature rapidly remodels in response to changes in blood flow. Because many biological systems are capable of robust error correction, it should be possible to design synthetic materials with similar functions. Error-correcting materials would revolutionize modern technologies, ranging from artificial joints that could be revitalized with error-correcting synthetic enzymes to last a lifetime instead of just a decade, to phone screens that repair themselves while recharging overnight. Creating these materials will require both fundamental discoveries in the physical and biological laws that govern these complex, dynamic interactions and innovations in computational and inverse design methods. So how do we design error-correcting materials without the luxury of billions of years of evolution? Come join us, we aim to learn how.
Complete list of work can also be found at my Google Scholar page.

Control over complex colloidal reactions with inverse designed components. Controlled disassembly of virus-like shells and, inspired by drug deliver applications, induced release of small particles trapped inside.
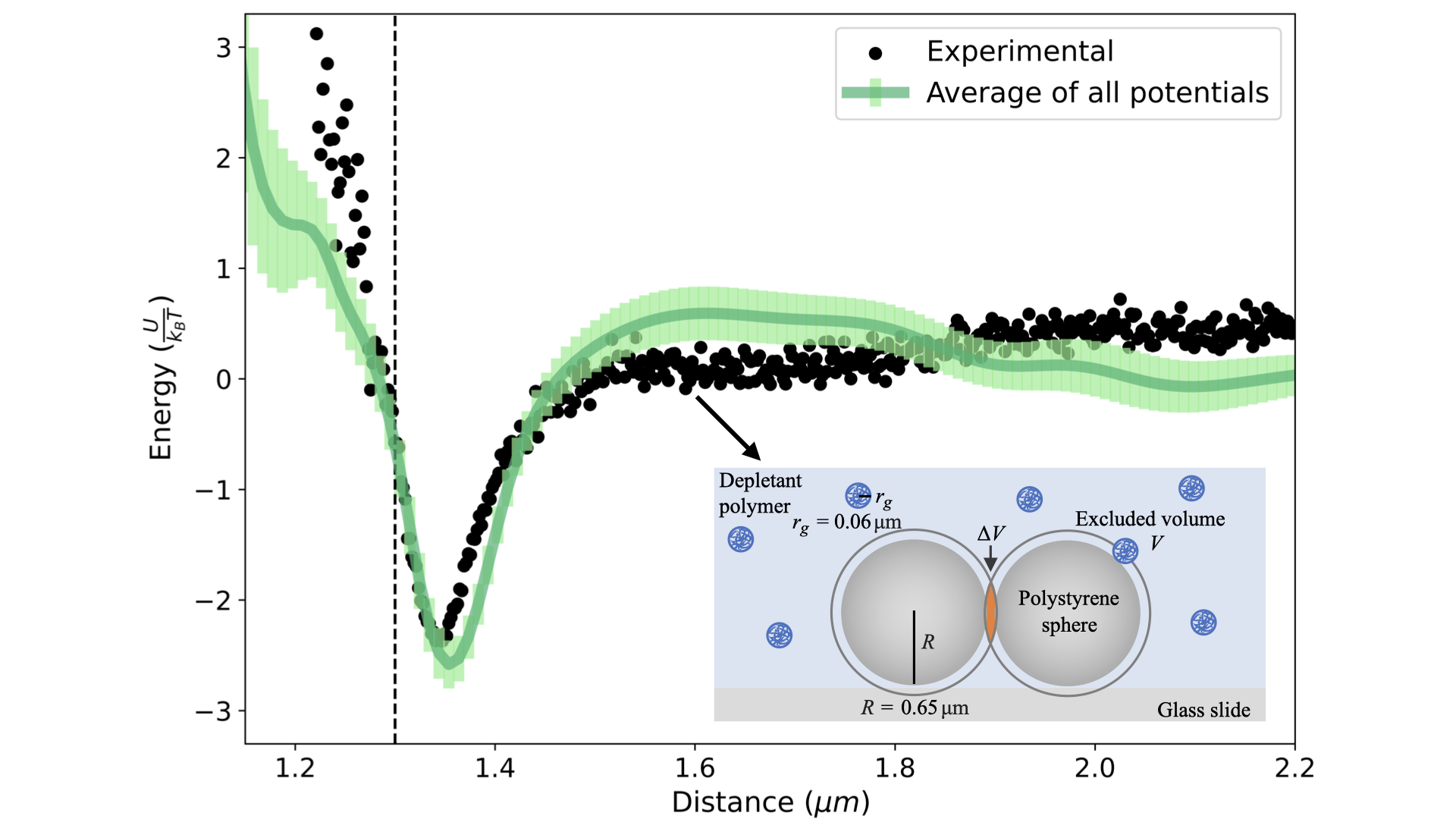
Novel method for extracting interactions from particle trajectories in and out of equilibrium. Validated on experimental colloidal data interacting via depletion forces.

Nonreciprocal wave-mediated interactions reveal a new form of activity that emerges as a collective property the system.

Computational design of anisotropic particle shapes that stabilize complex open lattices and enable self-limited assembly using automatic differentiation.
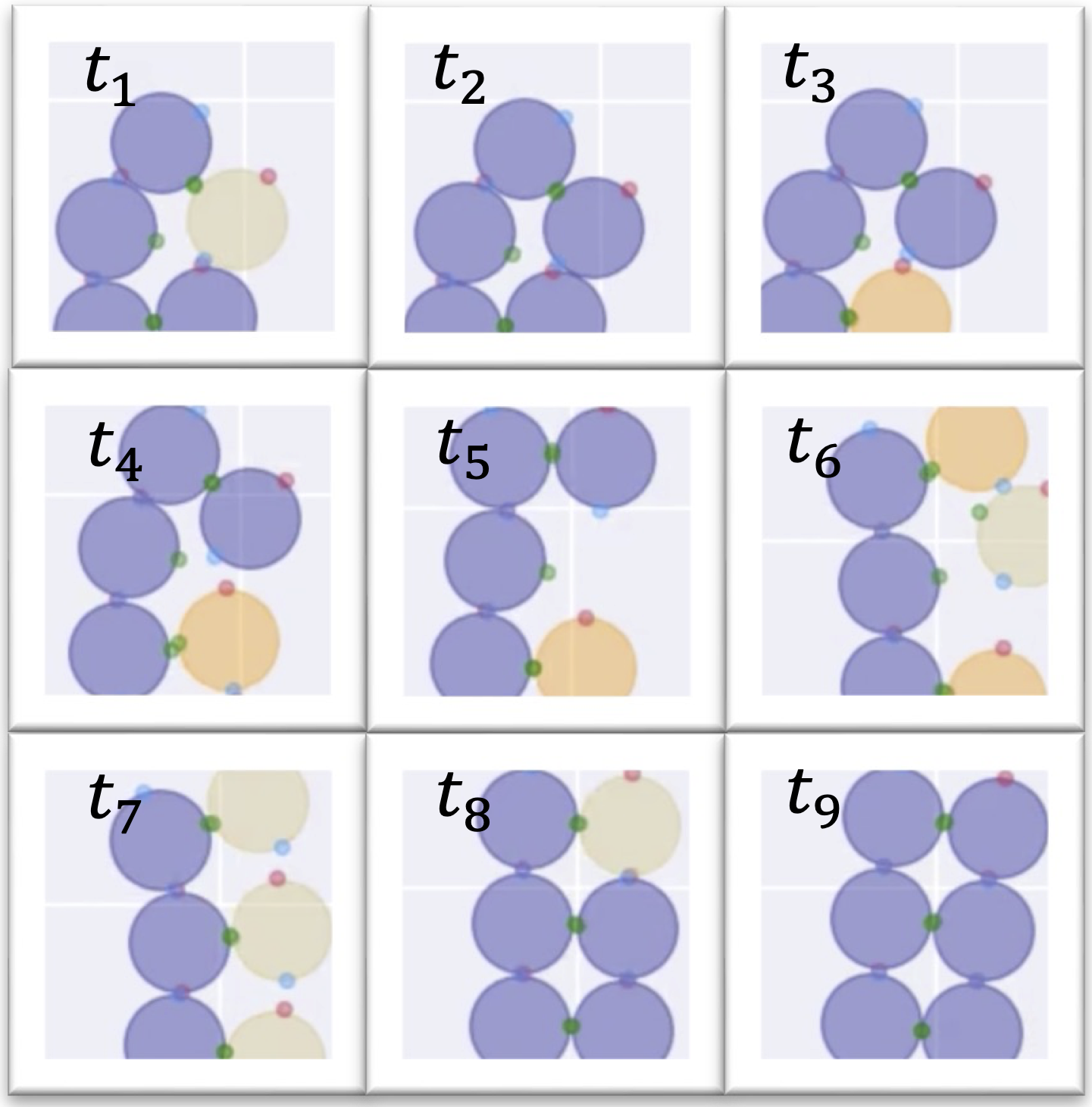
State-change based proofreading mechanism in colloidal particles undergoing self-assembly

Repeat expanded RNA, which is implicated in neurodegenerative diseases such as ALS, aggregates in vitro. Origin of this aggregation is presented, and a reentrant phase transition is observed as a result.
Introduces a particle tracking algorithm for correlated systems, such as dense suspensions or strongly interacting particles.
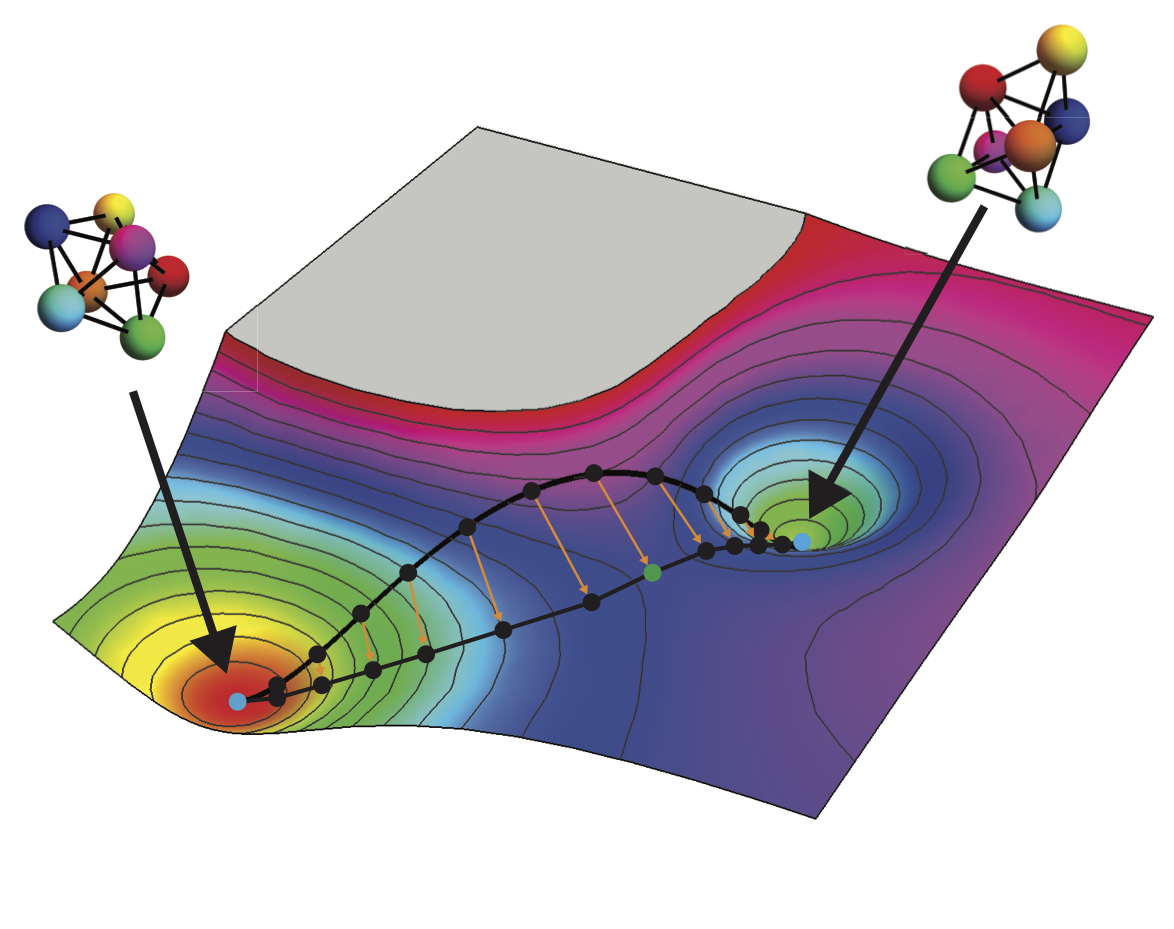
Inverse design of kinetic properties of self-assembled structures, including transition rates between small clusters and bulk crystallization rates.
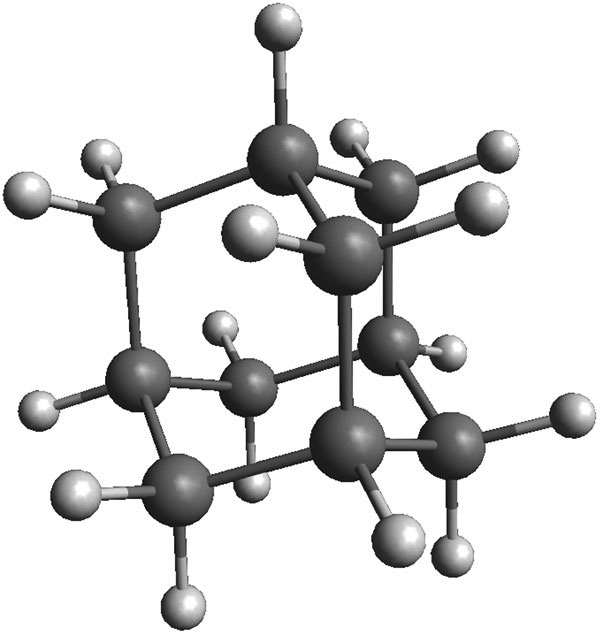
The rigidity of diamondoid molecules, and the resulting decrease in configurational entropy, drives their unique assembly properties.
Email: ella.king@northwestern.edu
Department of Physics, 726 Broadway, 8th floor, NYU, NY 10003 or Flatiron Institute, 7th floor, 162 5th Ave, New York, NY 10010.
You may also find me as volunteer mentor for the BioBus Junior Scientist Internship and as an occasional editor for WOWSTEM.